Reading FI data from an UFF file recorded from an Alpinion scanner
In this example we show how to read channel data from a UFF (Ultrasound File Format) file recorded with an Alpinion scanner. You will need an internet connection to download data.
by Ole Marius Hoel Rindal olemarius@olemarius.net and Muyinatu Lediju Bell mledijubell@jhu.edu 26.05.2017
Contents
Checking the file is in the path
To read data from a UFF file the first we need is, you guessed it, a UFF file. We check if it is on the current path and download it from the USTB websever.
clear all; close all; % data location url='http://ustb.no/datasets/'; % if not found downloaded from here local_path = [ustb_path(),'/data/']; % location of example data addpath(local_path); % We have two different Alpinion CPWC datasets, comment out the one to use filename='Alpinion_L3-8_FI_hyperechoic_scatterers.uff'; %filename='Alpinion_L3-8_FI_hypoechoic.uff'; % check if the file is available in the local path or downloads otherwise tools.download(filename, url, local_path);
Reading data
Now that the file is on the path let us create a UFF object to interact with the file. We open it in "append" mode.
uff_file=uff(filename)
uff_file =
uff with properties:
filename: 'Alpinion_L3-8_FI_hyperechoic_scatterers.uff'
version: 'v1.0.1'
mode: 'append'
verbose: 1
Let's first check if we are lucky and the file allready contains beamformed_data that we can display.
display=true; content = uff_file.index('/',display); has_b_data = false; for i = 1:length(content) if strcmp(content{i}.class,'uff.beamformed_data') has_b_data = true; % We found a beamformed data object! end end
UFF: Contents of Alpinion_L3-8_FI_hyperechoic_scatterers.uff at / - /b_data: b_data [uff.beamformed_data] size(1,1) - /channel_data: channel_data [uff.channel_data] size(1,1)
If the file had beamformed data, let's read that and the channeldata, before we can jump straight to displaying the data since it's allready beamformed
if has_b_data b_data=uff_file.read('/b_data'); channel_data=uff_file.read('/channel_data'); else
If it doesn't have any beamformed data at least it should have some channel_data. So let's read that.
channel_data=uff_file.read('/channel_data');
And then do the normal routine of defining the scan,
z_axis=linspace(1e-3,55e-3,512).';
sca=uff.linear_scan();
idx = 1;
for n=1:numel(channel_data.sequence)
sca(n)=uff.linear_scan(channel_data.sequence(n).source.x,z_axis);
end
setting up and running the beamforming
bmf=beamformer();
bmf.channel_data=channel_data;
bmf.scan=sca;
bmf.receive_apodization.window=uff.window.tukey25;
bmf.receive_apodization.f_number=1.7;
bmf.receive_apodization.apex.distance=Inf;
b_data=bmf.go({process.das_mex process.stack});
Now we can save this beamformed image to that file, so that we don't have to wait for the beamforming next time.
uff_file.write(b_data,'b_data');
end
UFF: reading b_data [uff.beamformed_data] UFF: reading sequence [uff.wave] Processed 256/256 UFF: reading channel_data [uff.channel_data] UFF: reading sequence [uff.wave] Processed 256/256
Display image
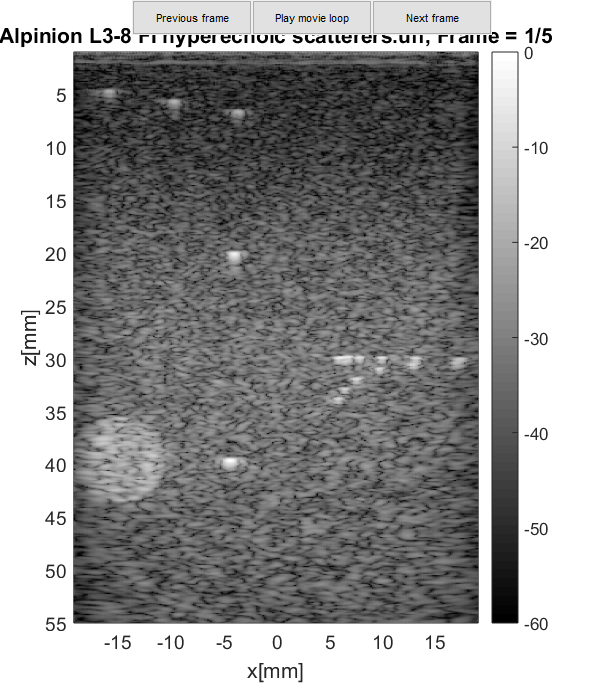
And finally display the image.
b_data.plot([],strrep(filename,'_',' '));
Write info about channel data
Let's look at the info given about this dataset
channel_data.print_info()
Name: FI dataset of hyperechoic cyst and points scatterers recorded on an Alpinion scanner with a L3-8 Probe from a CIRC General Purpose Ultrasound Phantom Reference: www.ultrasoundtoolbox.com Author(s): Ole Marius Hoel Rindal <olemarius@olemarius.no> Muyinatu Lediju Bell <mledijubell@jhu.edu> Version: 1.0.1